24 February, 2021
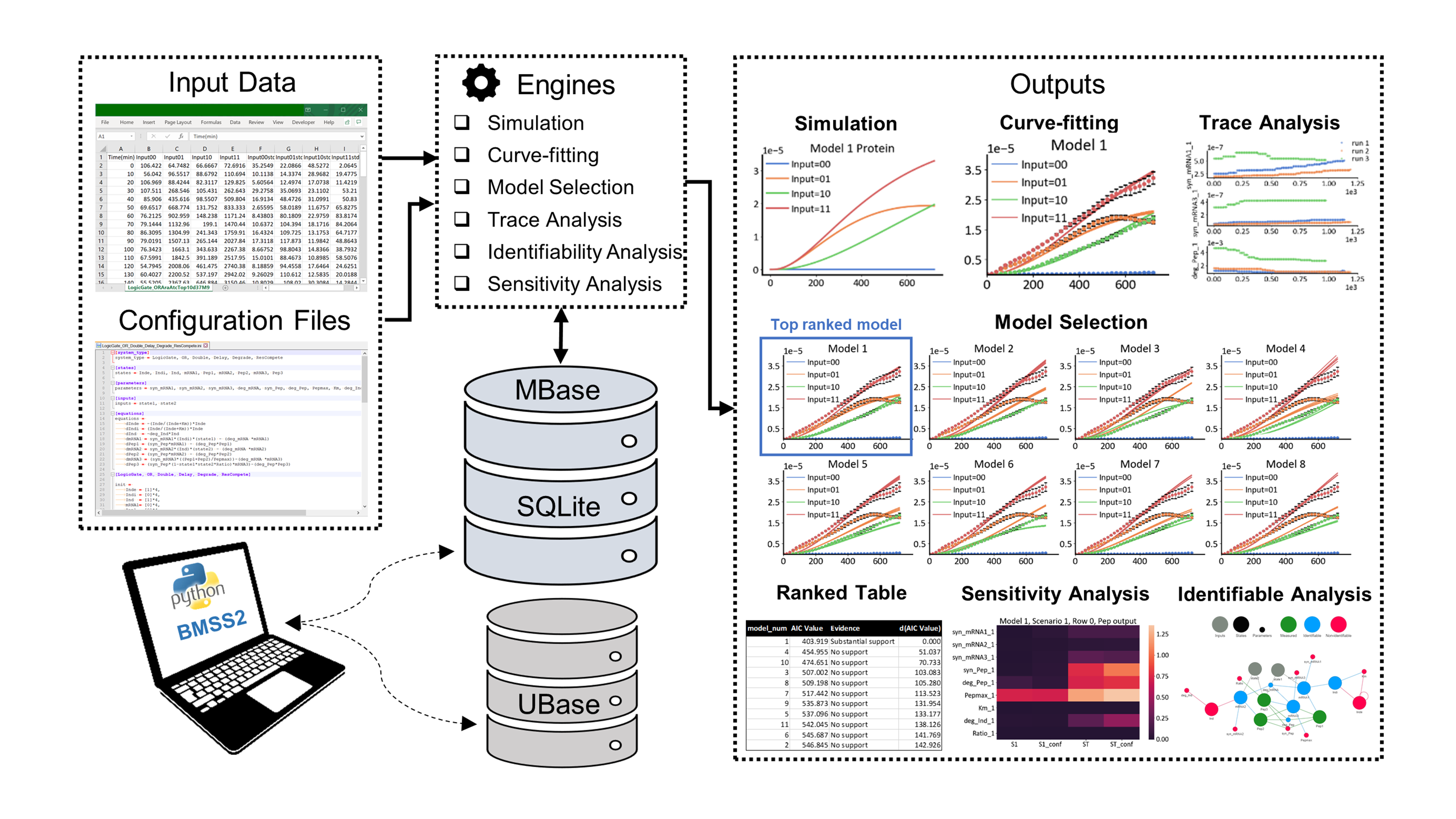
“BMSS2: a unified database-driven modelling tool for systematic model selection and identifiability analysis”
Our group just published the preprint:
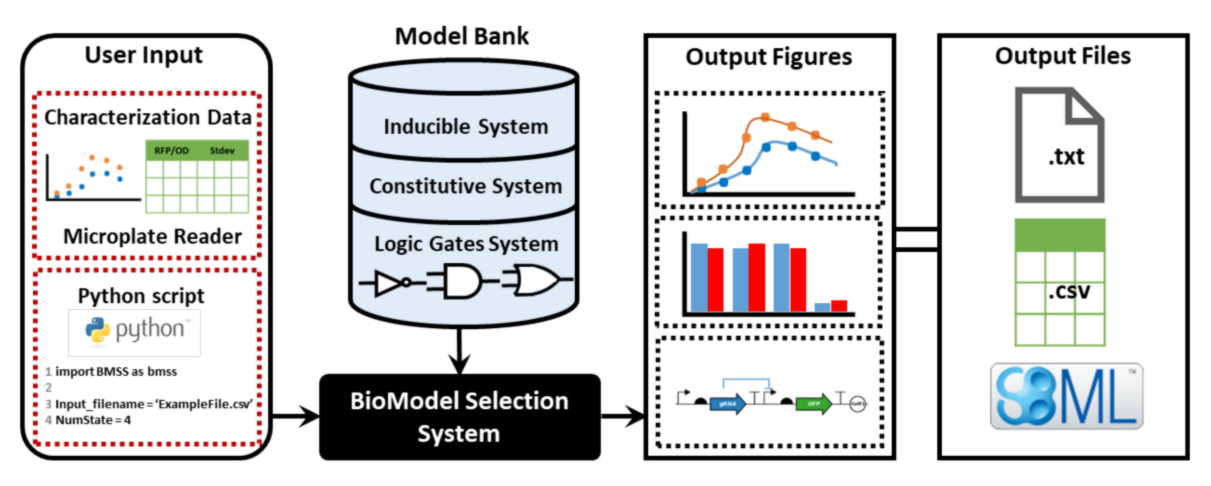
Modelling in Synthetic Biology constitutes a powerful tool in our continuous search for improved performance with rational Design-Build-Test-Learn approach. In particular, kinetic models unravel system dynamics, enabling system analysis for guiding experimental designs. However, a systematic yet modular pipeline that allows one to identify the “right” model and guide the experimental designs while tracing the entire model development and analysis is still lacking. Here, we introduce a unified python package, BMSS2, which offers the principal tools in model development and analysis—simulation, Bayesian parameter inference, global sensitivity analysis, with an emphasis on model selection, and a priori and a posteriori identifiability analysis. The whole package is database-driven to support interactive retrieving and storing of models for reusability. This allows ease of manipulation and deposition of models for the model selection and analysis process, thus enabling better utilization of models in guiding experimental designs.
11 October, 2020
New Force SCI webinar! 科研新势力, 科研共分享
Dr Zhang Jingyun has been invited to give a webinar in a Scientific Research Exchange Meeting (中国.新加坡)
Gene Editing and Synthetic Biology 关于基因剪辑知识 & 合成生物学
The talk features what is CRISPR, the development history of CRISPR, the key technology breakthrough, the patent war of CRISPR, application and future of CRISPR, ethical issues regarding the use of gene editing, and some of our lab works related to CRISPR. 什么是基因,什么是CRISPR, CRISPR发展简史,专利之争,技术应用,行业前景,伦理思考
26 September, 2020
Plasmid is available now on Addgene!
The link to Addgene: plasmid controlling motility in response to blue light
Zhang Jingyun, Luo Yuhuan and Poh Chueh Loo, Blue Light-Directed Cell Migration, Aggregation, and Patterning, Journal of Molecular Biology. First Online: 2 April 2020. DOI: 10.1016/j.jmb.2020.03.029
18 September, 2020
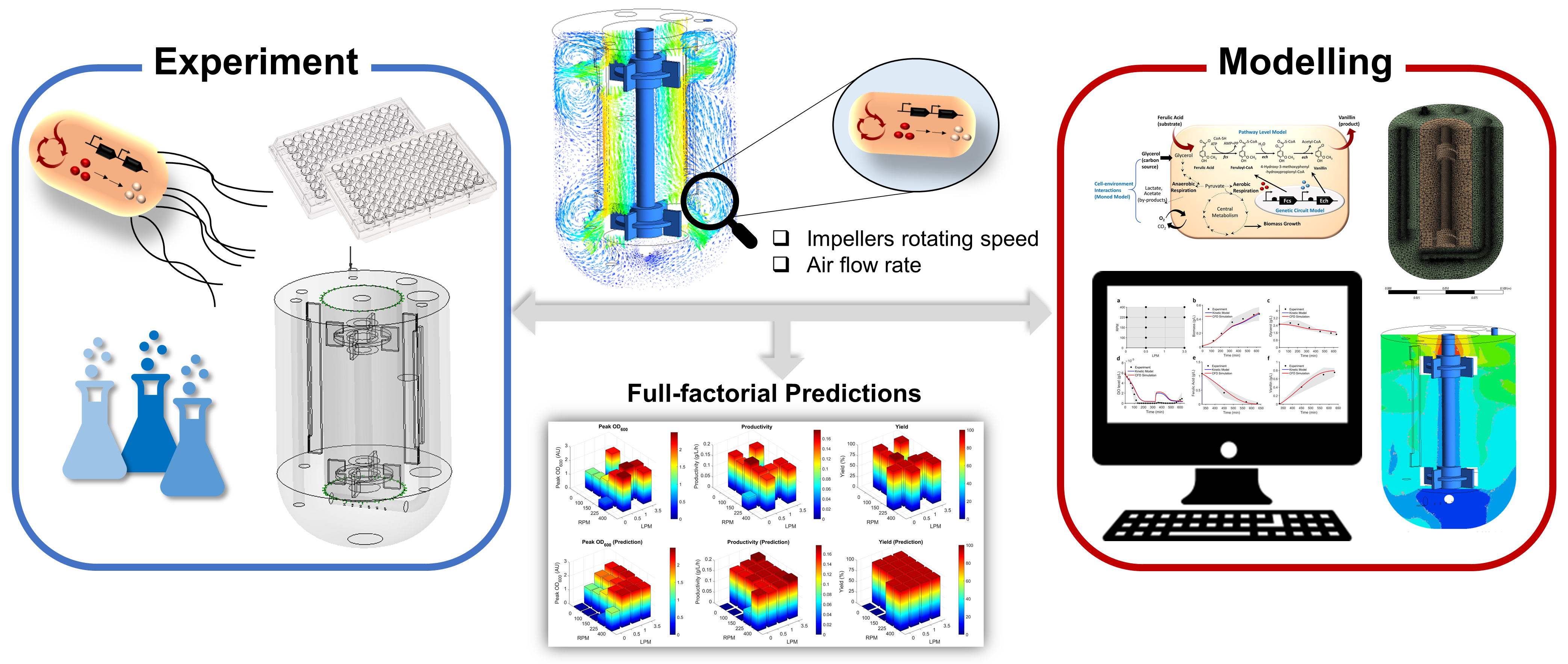
“A Model-driven Approach towards Rational Microbial Bioprocess Optimization.”
Our group just published the paper:
To enable a more rational bioprocess optimization, we have established a model-driven approach which systematically integrates experimental validation with modelling, executed from flask to bioreactor scale, using ferulic acid to vanillin bioconversion as a case study. The developed model covers multiple tempo-spatial scales of cellular reactions, ranging from gene expression kinetics, enzyme conversion, cell growth, oxygen uptake to oxygen transfer and mixing effects. The model well captured experimental profiles and full-factorial predictions were then performed to identify the desired operating conditions. Assisted with the 3D computational hydrodynamics of bioreactor, we were able to probe the mixing effects and metabolic heterogeneity of the cell factory. This model paves the way for a whole-cell, multiple scale analysis of gene expression, metabolic reactions and cell growth in the bioreactor.
2 April, 2020
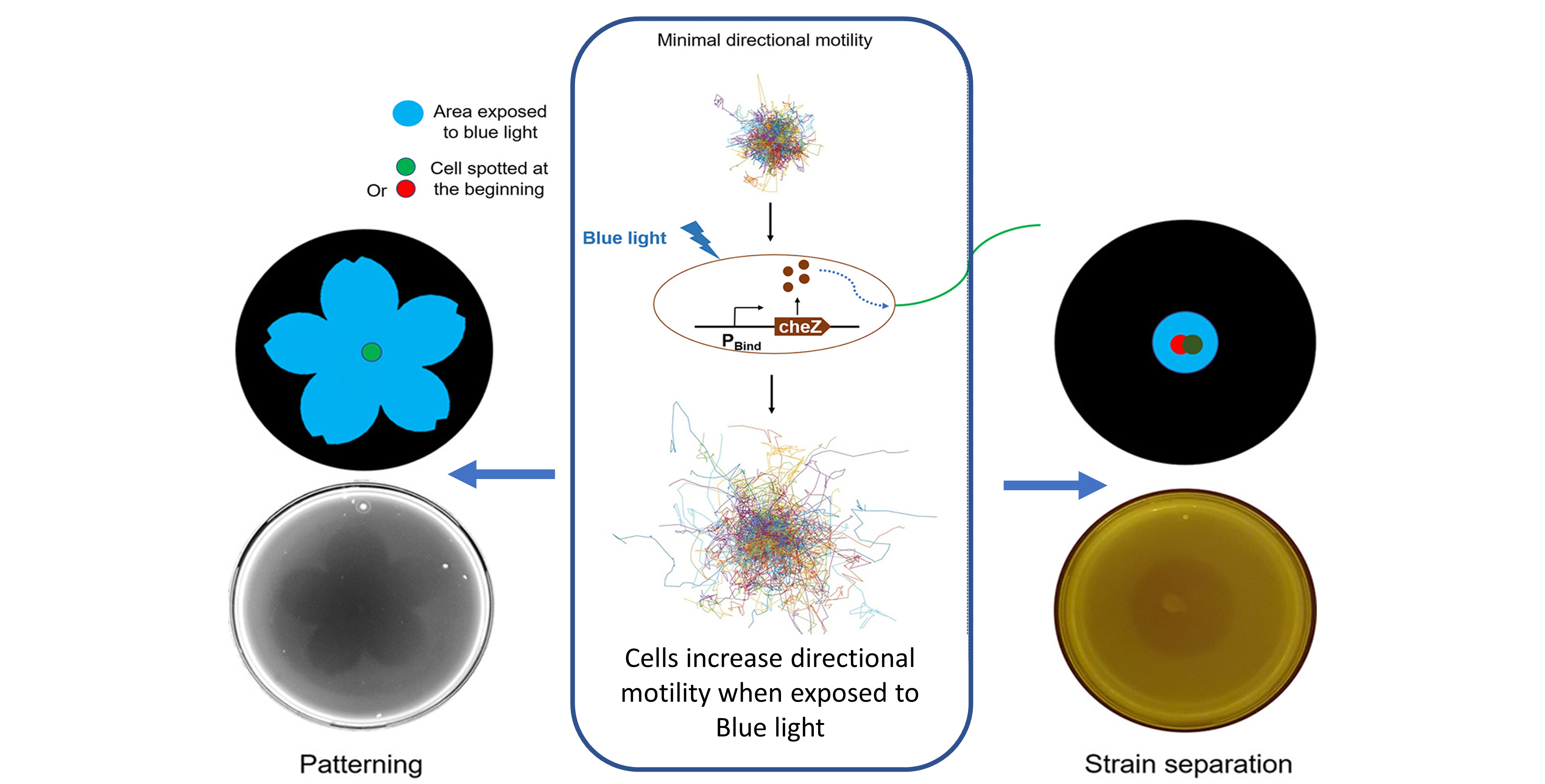
“Blue Light-Directed Cell Migration, Aggregation, and Patterning”
Our group just published the paper:
Zhang Jingyun, Luo Yuhuan and Poh Chueh Loo, Blue Light-Directed Cell Migration, Aggregation, and Patterning, Journal of Molecular Biology. First Online: 2 April 2020. DOI: 10.1016/j.jmb.2020.03.029
In this paper, we developed a blue light-regulated synthetic genetic circuit to control bacterial directional motility. As compared to chemically control motility, using blue light is non-invasive and is more flexible to create different prescribed patterns. The developed circuit enables the programmed Escherichia coli cells to directionally move away from the blue light (negative phototaxis). This also allows the control of cells to form aggregation with different complex patterns. Further, the blue light-inducible directional motility can also be used to separate two mixed strains.
3 December, 2019
Congratulations to Jingyun!
On 03 December 2019, Jingyun, our lab member, defended his PhD thesis, titled: “Programming microbes using synthetic biology approach to control biofilm development for continous bioproduction”. Congratulations!
29 November, 2019
Design of genetic circuits
Our group just published the book chapter:
This chapter aims to review the underpinning process involved in gene circuit design, with the emphasis on applying it to cell-based biosensors. Accordingly, appropriate computer-aided design tools to be used during the design and construction phases as well as modelling tools that facilitate the rational design-build-test-learn cycle will be explored. Lastly, a compilation of the common failure modes as faced by typical users and recommended potential engineering solutions is presented.
21 November, 2019
High capacity DNA storage scheme
Our group together with collaborators just published the paper:
Yixin Wang, Md Noor-A-Rahim, Jingyun Zhang, Erry Gunawan, Yong Liang Guan and Chueh Loo Poh. High capacity DNA data storage with variable-length Oligonucleotides using repeat accumulate code and hybrid mapping. Journal of Biological Engineering 13, 89 (2019).DOI: 10.1186/s13036-019-0211-2
To advance towards practical implementations of DNA storage, we proposed and tested a DNA data storage system enabling high potential mapping (bits to nucleotide conversion) scheme and low redundancy but highly efficient error correction code design. The advancement reported would move us closer to achieving a practical high-capacity DNA data storage system.
4 November, 2019
NUS iGEM team does it again! Wins Gold and more at the 2019 iGEM Giant Jamboree
Our NUS iGEM team has showcased their project, known as E.co LIVE, at the annual iGEM Giant Jamboree in Boston USA (31st Oct to 4th Nov 2019) with outstanding results! iGEM is the world largest annual synthetic biology student competition, with over 340 teams participating from universities around the world. This was NUS’s and Singapore’s best ever showing, where the team not only clinched a Gold Medal for the project but also went on to win two special awards – the Best Foundational Advance Project and Best Part Collection Awards. Apart from this they were also nominated for the Best Poster, Presentation, Wiki, Basic Part, Composite Part and Finalist awards. They were among the top 5 teams out of 194 undergraduate teams which participated in the competition!
This year the NUS iGEM team came up with the project to develop innovative solutions to enable the prolonged functionality of engineered bacteria, addressing the current bottleneck which limits the use of engineered bacteria to solving real-life problems, from cleaning up our environment to diagnosing diseases. Inspired by the phenomenon of hibernation, the students developed a “ON/OFF” growth switch to allow for fine control of the activity and energy consumption of the bacteria, extending their usage and thus bringing it closer to viability. Working hard throughout the year, the team demonstrated that the growth switch worked!
The team comprised of 9 highly motivated undergraduates from different faculties and disciplines, including Life Sciences, Biomedical Engineering, Chemical Engineering, Computer Engineering and Architecture, working closely to achieve this outstanding results. The team was mentored by NUS faculty, Prof Poh Chueh Loo (BME), Prof Jimmy Chih-Hsien Peng (ECE), Prof Matthew Chang (Biochemistry) and Prof Yew Wen Shan (Biochemistry), along with past ex-NUS iGEMers and members at SynCTI. From left to right in the picture: Joanne Lim, Suhasini Iyer, Cheng Kai Lim, Sheena Chan, Low Xi Zhi, Song Chun Yang, Zhao Hao, Prof Jimmy Peng and Prof Poh Chueh Loo.
We are very proud of our NUS 2019 iGEM team for their outstanding work and for being sterling ambassadors of NUS. For more information about their project, please click here.
If you are inspired and want to know more about NUS iGEM team, please contact A/Prof Poh Chueh Loo at [email protected].
9 May, 2019
Formal Launch of the Global Biofoundry Alliance
Professor Poh took part on the formal launch of the Global Biofoundry Alliance (GBA) in Kobe (Japan) on 09 May 2019. The GBA aims to increase coordination between its members in order to foster progress in the development of synthetic biology and associated areas.
More information can be found in the associated publication and in the GBA webpage.
29 April, 2019
The BMSS tool for automatic bio-modelling
Our group just published the paper:
BMSS is an automated bio-model selection system for gene circuit designs that supports bio-model automated fitting and selection processes, providing a means to efficiently derive the best model candidate that could capture the transient (time-series) dynamic profiles of a bio-part or device using characterization data. This system is an open source platform. The developed Python Package BMSSlib is available for downloading from GitHub. You can also find more information in our software page.
22 April, 2019
Codification method for data storage in DNA molecules
Our group together with collaborators just published the paper:
We propose a novel content-balanced run-length limited (C-RLL) code with an efficient code construction method, which generates short DNA sequences that satisfy both constraints at one time. Besides, we develop an encoding method to map binary data into long DNA sequences for DNA data storage, which ensures both local and global stability in terms of satisfying the biochemical constraints. The proposed encoding method has high effective code rate of 1.917 bits per nucleotide and low coding complexity.
25 October, 2018
Dual-input hybrid promoters lead to reconfigurable genetic circuits
Our group just published the paper:
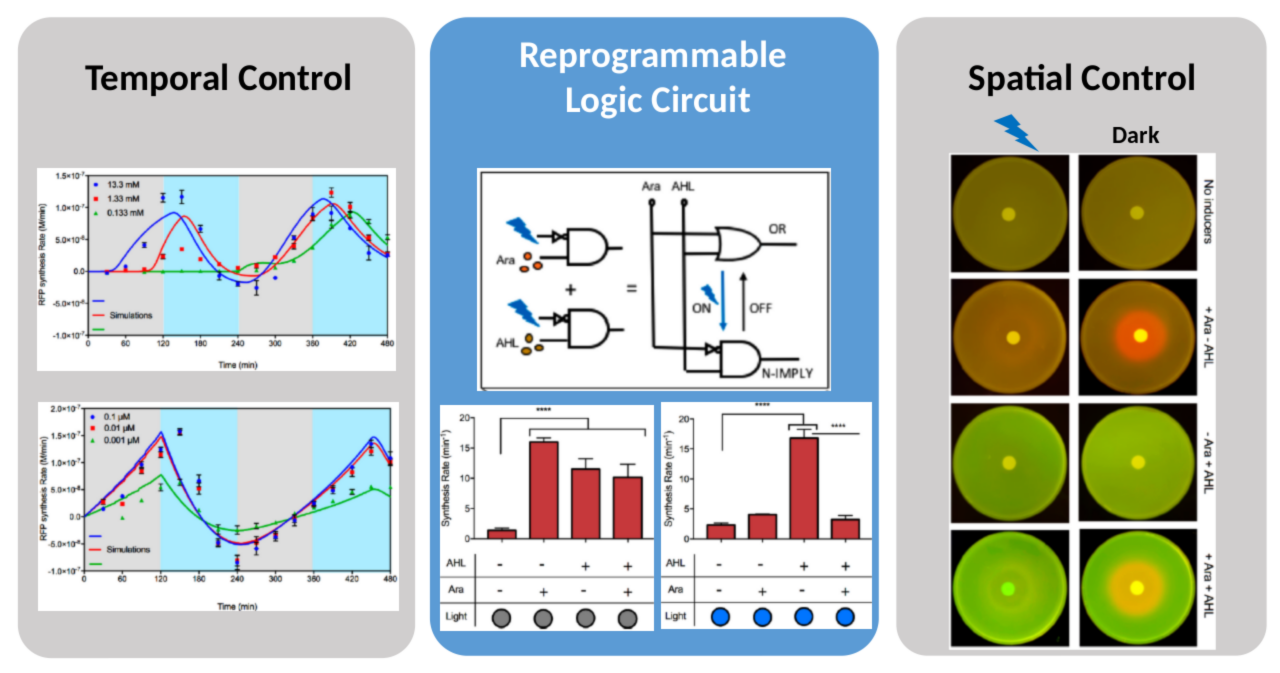
They built two dual-input hybrid promoters where both ligand and light inputs can be synchronously modulated to achieve desired amplitude, or independently regulated to generate desired frequency at a specific amplitude. This led to the synthesis of the first reprogrammable logic gene circuit capable of reconfiguring into logic OR and N-IMPLY logic on the fly in both space and time without the need to modify the circuit.
15-17 October, 2018
Dr. Teh Ai Ying presented her work in Shanghai
On 15-17 October 2018, Dr. Teh Ai Ying presented her work at the Synthetic Biology – Resource Allocation in Natural and Unnatural Systems conference. Check out her work aiming to improve the production of collagen for biomedical applications!
9 August, 2018
Biofilm engineering via wcaF regulation
Our group just published the paper:
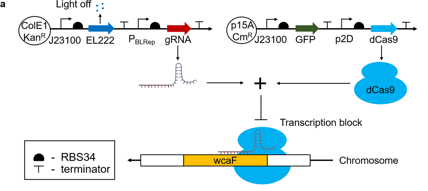
They developed synthetic gene circuits to control E. coli biofilm formation via the regulation of the gene wcaF, which is involved in the synthesis of colanic acid, a key polysaccharide in E. coli biofilm extracellular polymeric matrix. Combining CRISPRi/dCas9 and optogenetics, they achieved spatio-temporal control of wcaF, allowing to control biofilm thickness in time and space. Their results provide a novel technology with many applications in the field of biofilm engineering.